spora
Overview
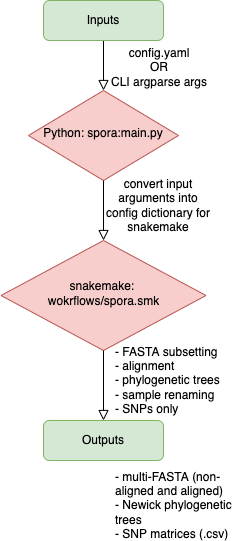
spora is a workflow written in snakemake and Python that aims to facilitate rapid generation of intermediate input files that are required for outbreak analysis for COVID-19 at PHO. The workflow is designed to be flexible with command line inputs, providing users with options that can be toggled depending on the nature of the outbreak request and the input files required for downstream outbreak analysis tools. At its core, spora is designed to accept only a small number of mandatory inputs from the user, and will use a standard set of bioinformatics tools to produce a number of output files such as alignments, trees, SNP matrices, etc., that are often the required inputs for downstream outbreak tools such as ggtree and/or civet/civet3.
Background
In a suspected outbreak, the epidemiology team should establish their initial search criteria; this is often described as the “case definition”. This includes an initial clinical description identifying the COVID-19 strain of interest (Single Nucleotide Polymorphisms (SNPs) of interest, genome of interest, etc), as well as restrictions on possible outbreak-affected patients based on “people, place, and time”. Based on this, the epidemiology team can supply WGS data from cases that meet this definition and are suspected to be a starting point in the analysis. These cases are then referred to as “focal cases”. When these are not provided at the outset, cases recorded through surveillance can be scrutinized against the epidemiological “case definition” to try and identify reasonable focal cases. At the same time, WGS data from COVID-19 cases known to not be involved in the suspected outbreak, based on criteria that exclude them from the case definition, can be used to define the background rate of disease. These “background cases” test whether the suspected outbreak is excess instances of disease.
Workflow